We will stop approving new registrations about an hour before each session. Please register in advance of that cutoff so you don’t get left out!
Session 4 Tutorial:
Computational Reproducibility Tools (Git/Github, Conda, Docker/Singularity containers)
This page contains all the info you need to participate in Session 4 of the SCINet Geospatial Workshop 2020.
A note about running this tutorial in your home directory: some participants got errors either downloading to or running the tutorial from their home directory because they did not have enough space free in their home directory. If this happens to you, download to and run the tutorial from a project directory. If you are a new HPC user and don’t yet have a project directory, you can request a small increase in space to your home directory from the SCINet VRSC at scinet_vrsc@usda.gov.
The session recording is available for anyone with a usda.gov email address and eAuthentication at (location coming soon).
Learning Goals:
Git/Github
- understand the basics of how Git/Github works and the benefits of using them to version and archive your codes
- create Github repos using two different workflows
- understand how to use git fork/clone/add/commit/push and make a pull request
Conda
- understand the basics of how the Conda package/environment management system works and the benefits of using it
- use Conda to create environments and install software on the Ceres HPC system
- export your Conda environment details so you can quickly recreate your software environment for any project
Containers
- understand the basics of containers and the benefits of using them for code portability and reproducibility
- navigate Dockerhub, build a Singularity container on Ceres from a Docker image, create a Docker image, create a DOI for a container image
Contents
- Why Use Conda?
- Terminology
- Accessing Conda on Ceres
- Using Conda Environments to Create Isolated Software Workspaces
Containers (Docker and Singularity)
- Why Use Containers?
- Terminology
- Finding an Existing Docker Image
- Using a Docker Image on Ceres with Singularity
- Building a Docker Image
- Example: GitHub/Zenodo + Docker Hub Setup
Session Rules
GREEN LIGHT, RED LIGHT - Use the Zoom participant feedback indicators to show us if you are following along successfully as well as when you need help. To access participant feed back, click on the “Participants” icon to open the participants pane/window. Click the green “yes” to indicate that you are following along successfully, click the red “no” to indicate when you need help. Ideally, you will have either the red or green indicator displayed for yourself throughout the entire tutorial. We will pause every so often to work through solutions for participants displaying a red light.
CHAT QUESTIONS/COMMENTS TAKE FIRST PRIORITY - Chat your questions/comments either to everyone (preferred) or to the chat moderator (Rowan or Kerrie - whoever is not actively presenting) privately to have your question/comment read out loud anonamously. We will address chat questions/comments first and call on people who have written in the chat before we take questions from raised hands.
SHARE YOUR VIDEO WHEN SPEAKING - If your internet plan/connectivity allows, please share your video when speaking.
KEEP YOURSELF ON MUTE - Please mute yourself unless you are called on.
Git and Github
Why Use Git/Github?
“Git tracks the changes you make to files, so you have a record of what has been done, and you can revert to specific versions should you ever need to. Git also makes collaboration easier, allowing changes by multiple people to all be merged into one source. So regardless of whether you write code that only you will see, or work as part of a team, Git will be useful for you.”
Git and Github may seem excessively complicated, but that is because these tools were created for major software development projects. As research scientists, we can use Git and Github as tools for version control, online back up of our files, collaboration/sharing, and reproducibility by only learning a handful of commands which we will introduce in this tutorial. Once you have these handful of commands down you’ll be on your way. Leave the complicated stuff for the software developers to worry about!
Version Control
Git is a powerful tool for keeping track of edits to your files over time (version control). Using git eliminates the process of saving duplicate codes (e.g., project-code.py, project-code-rev1.py, project-code-rev2.py, project-code-test-this-or-that.py, project-code-latest.py, etc.). We’ve all done this type of insufficient versioning of our codes and thoroughly confused ourselves or lost track of which code contains which important changes, etc. Git solves this problem. Instead of creating multiple files, Git keeps track of your file changes in a much tidier way by attaching a change message called a “commit” each time you make a major change to your files. A commit is like a snapshot of your code at a point in time and you can revert back to that snapshot any time you want. Git also has a structure called “branches” that allows you to add/test new bits of experimental code inside an existing code you’ve developed without affecting your existing code file.
Backups
Github, being an online repository, can be used as a backup for the codes (and more) on your local computer. By pushing your work to Github every so often, you can create an online backup of your work. As a bonus, this also means you’ll be able to access your codes (and more) from anywhere because they are online.
Collaboration and Sharing
Git and Github are amazing tools for collaborating on research computing. Collaborators can simultaneously code locally on their own machines, push their file changes to Github, and Github will discover any conflicts and help you merge all the changes together. Also, pushing your local code to Github allows you to give out a URL to anyone you want to share your codes (and more) with. Note: you can use Github for version control of manuscripts and figures, but you do lose some functionality when you are tracking something other than codes, markdown files, or html.
Reproducibility
Git and Github greatly improve computational reproducibility by enabling others access to your scientific codes (if you choose) and history of file changes. Github also interfaces with other computational reproducibility tools like Docker/Dockerhub for creating/sharing isolated software environments called containers. More on containers in the next segment.
Git Terminology
GIT - version control software that you use at the command line to keep track of edits to your scientific codes over time.
GITHUB - an online repository hosting service where you can archive your scientific codes online. You can create a free personal Github account where you can archive your scientific codes into different “repositories” within your account. If you keep your Github repos up to date, it also allows you (and collaborators) to access your codes from anywhere.
REPOSITORY (REPO) - A directory or storage space where your projects can live. A repo can be a local folder on your laptop/HPC system (local repo) or it can be a storage space on GitHub (online/remote repo). Inside a repo is where you keep your code files, text files, etc. Generally, each of your local repos is also associated with at least one online Github repo called a “remote” repo.
CLONE - the process of copying an entire existing Github repo to your computer (copying to a local repo)
FORK - the process of copying an entire existing Github repo to your own Github account (copying to an online repo)
BRANCH - each branch in a repo is an isolated space where you can work/develop your codes without affecting the work saved in other branches. Each repository has one default branch called Master (soon to be renamed Main) and can have multiple other branches. A new branch is always created from an existing branch. The following example is one way to use branches effectively. If you have a functioning code on the Master branch and you want to experiment with code changes without affecting your functioning code on Master, you can create a new branch called Dev, for example. You could then make changes to your code on the Dev branch which would be entirely separate from your code on the Master branch. If you like your changes, you could incorporate them into your code on the Master branch by merging the Dev branch into the Master branch.
COMMIT - the process of saving a “snapshot” or versions of your files. Similar to saving a file that’s been edited, a commit records your changes to one or more files on a branch. Git assigns each commit a unique ID that indentifies the specific changes, when the changes were made, and who created the changes. When you make a commit, you also attached a short description of the changes you made. It’s wise to group similar edits together and include them in a single commit and also to commit your changes every so often so that you have a good number of “snapshots” available to you in case you break your code and need to look back to earlier versions. For example, you could code all day and do one commit at the end of the day and have one snapshot/version of your code, but it may be more useful to commit more often that way you will have multiple snapshots of your work throughout the day that you could revert back to if necessary.
STAGE - the process of “telling Git” which edited files should be included in each commit that you make. Just because you change a file in your repo does not mean that it will be automatically included in your repo snapshot when you commit. You must first “stage” the edited files so that Git knows how to make the snapshot.
PULL - the process of updating your local repo to include changes made to the remote (online) repo. You “pull” down any edits from the online repo and merge them into your local repo.
PUSH - the process of updating your online remote repo with the changes you have made to your local repo. You “push” up any edits from your local repo and merge them into your online remote repo.
PULL REQUEST - the process of requesting that changes you have made to your repo branch be incorportated into someone else’s version of the repo. For example, you fork (copy) someone’s existing Github repo to a repo on your own Github account and you then make changes to some of the files. If you want the original repo that you copied from to incorporate your changes into their repo, then you would make a pull request. It is called a pull request because you are requesting that someone else pulls and merges your edits into their online repo.
MERGE - the process of combining multiple sequences of commits into one unified history. Most often used to incorporate changes from one branch into another. A merge happens when you pull, push, and pull request.
Using Git and Github to Archive and Version Your Codes
A Workflow Starting From Github
This example workflow demonstrates how to:
- copy an existing Github repo to your own Github account (Step 1),
- make a local copy of your Github repo on your computer, in this case your Ceres account (Step 2)
- make changes to the repo files locally (Step 3),
- push your local repo changes to your Github repo (Step 4), and
- have your repo edits incorporated back into the online repo that you originally copied from on Github (Step 5)
Step 1: Fork a Github Repo (on Github, copy an existing Github repo to your own Github account)
a. Login to Github with your username and password.
b. Find the Github repo that you want to copy. Let’s copy the repo called “Spoon-Knife” on Kerrie Geil’s Github page.
- In the black Github search bar at the top left where it says “Search or jump to”, type kerriegeil, and select the dropdown list option that says search kerriegeil All Github.
- On the bottom of the resulting menu click Users, then click Kerrie Geil on the right to see all the public repos on her account.
- Click on the Spoon-Knife repo. You should be here: https://github.com/kerriegeil/Spoon-Knife.
c. Click Fork on the top right of the repo page and then click on your github username when you are asked where to fork to. You have now copied the repo to your own account. Notice the URL you are at- it should be github.com/yourusername/reponame
Step 2: Create a Local Repo on Ceres From Your Github Repo (git clone)
a. SSH into your Ceres account from a terminal/command line. See the Ceres Quick Start Guide for instructions.
b. Navigate to the location on Ceres where you want to copy the repo. It’s small so putting it in your home directory is fine (~/home/username).
c. Go back to Github to get the URL of your Github repo. On your repo page, click the green Code button to find the repo address. It should be in the form github.com/yourusername/Spoon-Knife.git
d. Back at the command line type the following to make a local copy of your Github repo
git clone paste_or_type_the_full_repo_URL_including_https://
Step 3: Make Changes to Your Local Repo and Make Multiple Commits (git add, git commit)
a. Create some new text files using your name as the file name
cd Spoon-Knife
touch yourname1.txt yourname2.txt yourname3.txt
ls
b. Check the status of your edits. The changed files should appear as untracked files
git status
c. Stage the edits before committing. You are “telling Git” what files you want to include in a future commit.
git add yourname1.txt
git add yourname2.txt yourname3.txt
d. Check the status of your edits. The changed files should now appear as changes to be committed
git status
e. Commit the edits to include them in a snapshot of your updated local repo
git commit -m "create new text files"
f. Check the status of your edits again. The changed files should no longer appear in the status output. If you’ve committed all your changes you should see “nothing to commit, working directory clean”
git status
g. Let’s do it again. Edit the file hi-my-name-is.txt to include your name
nano hi-my-name-is.txt
then type your name, hit Ctl+O then enter to write, Ctl+X to exit
h. Stage your edits
git add hi-my-name-is.txt
f. Another commit
git commit -m "update with my name"
g. Check the status of the changes. Working directory should now be clean, meaning all changes have been committed to an updated “snapshot”
git status
Step 4: Push Your Local Changes Up to Your Remote Repo on Your Github Account (git push)
a. If you want to double check what remote repo(s) is/are associated with your local repo, at the command line type
git remote -v
You should see that your remote repo is called “origin” for short and that the URL to the remote repo is github.com/yourusername/Spoon-Knife.git, the same URL we used when we cloned the repo to your local computer
b. Push your local changes to your remote repo on your Github account with
git push -u origin master
Enter your Github username and password when you are asked for it.
c. Go back to your repo on Github and refresh the page. You should see your new files appear now on Github
Step 5: Make a Pull Request (request that your changes be incorporated into the original repo)
a. To add your repo changes into the original repo at kerriegeil/Spoon-Knife, navigate to your yourusername/Spoon Knife repo on Github.
b. Now submit a pull request. You are requesting that kerriegeil incorporate (or “pull”) your edits into her repo
- click the Pull Requests tab
- click the green New Pull Request button on the right
- ensure the base repository is kerriegeil/Spoon-Knife and the base branch is Master (left side of the arrow)
- ensure that the head repository is yourusername/Spoon-Knife and the compare branch is Master (right side of the arrow)
- click the green Create Pull Request button and enter a title for your pull request
- the fate of your pull request now lies with the owner of the original repo. They will see your pull request and approve/merge your changes into their repo
A Workflow Starting From the Command Line
Sometimes you’ll want to start a brand new local repo right from the command line and then push your local repo up to Github. Here’s how to do it.
Step 1: Initialize a New Local Git Repo
a. Create a new local directory where you want your new local repo to live and navigate into it
cd
mkdir my_new_repo
cd my_new_repo
b. Initialize a new local git repository
git init
Step 2: Edit away!
Adding content to your local repo is the exact same process as Step 3 in the previous workflow
touch newfile.txt
git add newfile.txt
git commit -m "create new text file"
Step 3: Create a Remote Repo on Github and Push Your Local Repo Up to Your Github Repo
a. Login to your Github account
b. Navigate to your repositories page (click the Repositories tab or on your user profile dropdown menu select “your repositories”), click the green New button
c. Give your new Github repo the same name my_new_repo
d. You can skip the section about creating a readme, license, gitignore because you’ll be importing an existing repo
e. Follow the instructions to push an existing repo from the command line
git remote add origin https://github.com/yourusername/my_new_repo.git
git push -u origin master
Enter your Github username and password when you are asked for it.
f. Refresh your Github repo page, you should see all your local files now in your Github repo
Additional Git Commands
remote repos: origin vs. upstream
Upstream generally refers to the original repo on Github that you have forked from. Origin generally refers to your fork of the repo on your Github.
When you fork a repo on Github, you are making a copy of the repo to your own Github account. When you clone one of your Github repos to your local computer the default remote repo “origin” is set to the location of your remote repo on your Github. You would use git push and git pull to upload/download changes between your local repo and your Github repo.
To keep track of the repo you forked from originally you will have to add an additional remote repo called “upstream”. Then you will be able to pull down changes that have been made to the original repo using git fetch.
git remote add upstream https://github.com/username_who_you_copied_from/their_repo_name.git
See your remote origin and upstream repositories with:
git remote -v
keep your local and remote repos in sync
Even if you aren’t collaborating with someone, it’s a good idea to make sure your local repo and your repo on Github are in sync with each other each time you start and finish working. This way you can avoid merge conflicts later. Make sure your remote origin is set correctly as described in the remote repos section above.
Push up your local changes to your Github repo every time you finish working locally:
git push
Pull down any changes that have been made to your remote Github repo when you start working locally:
git pull
This is especially important if you are working with a collaborator on a repo. You’ll want to incorporate any changes your collaborator has made to the repo before you start working. If the original repo you are collaborating on was created in your own Github account, use the commands above.
If the original repo was created on your collaborator’s Github account and you forked it, you’ll have to add their repo as remote upstream (described in the remote repos section above). Then to pull down changes from their repo to your local repo:
git fetch upstream
access repo commit history
There are two ways to see every available snapshot of your repo (history of commits). On Github in your repo, click the commits link located right under the green Code button. From the command line:
git log
We won’t cover how to revert your repo to a previous commit in this tutorial but you should know that commit number you see on Github commits or in the git log output at the command line is what you need to access an earlier version of your repo.
create and switch to a new branch
If you need a new isolated area to experiment with changes to your code without affecting your codes on the deafault Master branch, create a new branch and switch to it with
git checkout -b new_branch_name
To show all your branches and which branch you are currently on:
git branch
To switch back to your Master branch:
git checkout master
Note: from the command line creating a new branch does not create a new folder in your repo directory. Be careful to always be aware of which branch you are working on- from the command line it is not obvious.
Anaconda (Conda)
Why Use Conda?
Conda is not only a tool that allows Ceres users to install software themselves on the HPC system, but it is awesome for reproducibility.
Conda allows you to keep track of exactly what software (and all of the many dependencies) you are using to execute your scientific codes. This is very handy to know, especially if a software package update ends up breaking your code! You are able to save your entire software environment and recreate it any time you want if something goes wrong.
Conda also let’s you run separate instances of the same software, for example, maybe you have one project where you use Python 2 and another where you use Python 3. Conda allows you to make isolated software environments for your projects so you can run the software you need for each project without conflicts.
Conda Terminology
CONDA - an open source software package management and environment management system that runs on Windows, Mac, and Linux. Conda can find, install, run, and update software packages and their dependencies as well as create, save, and load software environments on your computer. It was created for Python programs, but it can package and distribute software for any language.
PACKAGES - pieces of software and their dependencies
DEPENDENCIES - pieces of software that are require for other software to run successfully
ENVIRONMENT - a conda software environment is a directory that contains a specific collection of conda packages that you have installed. It is isolated from your other conda environments such that when you change one environment, your other environments are not affected.
ACTIVATE - the process of “entering” a specific conda environment. When you activate an environment you are telling Conda which isolated set of software packages to use to execute your codes.
SPECIFICATION FILE - a text file that lists every single package that’s in a specific environment, including the packages that are specific to the operating system you are running on. A specfile can be used to recreate an environment. The file tells you what operating system you can recreate the environment on.
YML - another type of file that contains information that can be used to recreate an environment. Operating system-specific packages can be excluded from a yml file in order to recreate similar environments across different operating systems.
Accessing Conda on Ceres
Conda is available on Ceres without users having to install it themselves. Actually, it’s recommended that users NOT install Conda themselves. Conda can be accessed through the software module system or by logging into the system using JupyterHub as described below. See also the Guide to User-Installed Software on Ceres with Conda on the SCINet website.
From the Module System
After SSHing into Ceres, it is easy to load Conda from the software module system.
module load miniconda
At the time of this writing, the default Conda on Ceres is miniconda/4.7.12.
You will then want to immediately issue the following command which will put you in the base environment:
source activate
Note: If you forget to source activate and later try to conda activate my_env, you will get a command not found error and will be instructed to conda init. Despite the standard output instructions, DO NOT EVER TYPE conda init ON THE CERES HPC. It will make a permanent modification to your $PATH that doesn’t play nice with the software module system or with Jupyter. If you accidentally conda init you will have to modify your .bashrc file to remove any conda initialization info. See the section of the Guide to User-Installed Software on Ceres with Conda highlighted in red for more detail about how to fix your .bashrc in this case.
After you source activate and are placed in the base environment, you will then be able to conda activate my_env with no problems.
From JupyterHub
When using JupyterHub to login to Ceres you will also have access to Conda.
JupyterHub login with no container
If you login to Ceres with JupyterHub and are not using a container, you will automatically have access to Conda- no need to load the module. At the time of this writing the default is miniconda/3.6.
always remember to source activate immediately and to never conda init
see the above From the Module System for more detail
The Conda version shouldn’t really matter, but if you run into problems with this older version you can always open a terminal in JupyterLab and execute the same commands as in the above section From the Module System to load a more up-to-date Conda version from the software module system.
JupyterHub login with a container
If you login to Ceres with the workshop image “data_science_im_rs_vSCINetGeoWS_2020.sif” you will have access to miniconda/4.8.3.
always remember to source activate immediately and to never conda init
see above From the Module System for more detail
Using Conda Environments to Create Isolated Software Workspaces
The best practice for using Conda is to never install packages in the base environment.
One reason for this is because the base environment is a dynamic space that changes with Conda version updates whereas your other environments are isolated. You should create at least one other environment to install packages in. A better practice is to create a separate environment for each one of your projects. It’s much easier to update certain software packages in your environments if your projects each have their own “sandbox”. In addition, on Ceres you’ll want to save the environments you create in your home directory or in your project /KEEP directory, whereas the base environment lives outside of your personal user account.
Many of the environments that we as geospatial researchers build contain enough software packages that the environment will take up multiple GBs of storage space. Because our environments are generally large, they take a while to build. The best practice for creating Conda environments on Ceres is to first open an interactive compute session (i.e. don’t create/remove environments or install packages on the login node).
At this point you should be logged in to your Ceres home directory preferably by using JupyterHub (with no container) and have opened a terminal in JupyterLab. You could also SSH in and load the miniconda module as described in the From the Module System section above, but the remainder of this Conda tutorial will assume you are logged in from JupyterHub.
The Conda Basics
Create an New Environment
In this tutorial we will create an environment that you can run the Session 3 Tutorial with in JupyterLab.
To open an interactive compute session:
salloc
You are now on a compute node as opposed to the login node.
To get into the base Conda environment:
source activate
Create a new environment with conda create --name environment_name package1 package2 package3.
conda create --name session3_env python=3.7 numpy dask dask-jobqueue
Make sure you hit enter when Conda asks if you want to proceed. This build will likely take about 5 minutes.
View all Environments
When the build is finished and your command prompt returns, view all your environments with:
conda env list
Activate an Environment and Install More Software
To activate your new environment
conda activate session3_env
To access this environment in JupyterLab you will need to install the ipykernel package. We could have done this with our conda create but are doing it after the fact to demonstrate how to add additional packages into an existing environment. Make sure your session3_env is activated and
conda install ipykernel -y
Notice how the -y allows you to bypass the “proceed ([y]/n)?”
View Software in an Environment
To view all the packages in the active environment
conda list
Change or Deactivate an Environment
To change environments
conda activate base
will switch you to another environment, in this case the base environment.
Or,
conda deactivate
will switch you from a non-base environment back to whichever environment you were in previously.
Export the Environment Software List to a Specification File
Create a specification file that lists every single software package that’s in an environment, including the packages that are specific to the operating system you are running on. You can save this file wherever you want. Here we’ll save it in the Conda envs folder where the rest of your environment information lives.
conda activate session3_env
conda list --explicit > ~/.conda/envs/session3_env_spec_file.txt
View the file you just made
cat ~/.conda/envs/session3_env_spec_file.txt
Notice at the very top of the file there are instructions for recreating the environment from this file, we’ll cover this shortly. It also tells you what operating system you can recreate the environment on, in our case you can recreate this environment on 64-bit linux platforms. Recreating the environment on other platforms will likely not work.
Export the Environment Across OS Platforms
To use an environment across different operating systems you need create a yml file with a list of software that excludes all the OS-specific software in the environment and only includes the major software packages (without all the dependencies). For our session3_env this means the packages we explicitly installed: python 3.7, numpy, dask, dask-jobqueue, ipykernel.
conda env export --from-history > ~/.conda/envs/session3_env.yml
View the file you just made
cat ~/.conda/envs/session3_env.yml
As with the specification file we made earlier, you can use this yml file to recreate your environment on the same machine. You can also use a yml to create a similar environment on another machine running a different operating system. This means you can share this yml with other scientists too. Note if there is a –prefix line in your yml file you’ll need to modify that if recreating on a different machine or in a different file structure (e.g. different Ceres user). Note: this isn’t a 100% foolproof method of getting your codes to run across different platforms because some software just isn’t fully supported on all operating systems. Being able to run your codes successully across operating systems is the major benefit of using containers, which we will cover in the next segment.
Delete an Environment and Recreate from a Specification File
Let’s say you run out of space to store all your environments. No problem, as long as you have a specification file that lists the environment software, you can go ahead and delete your environment because you’ll be able to recreate it anytime.
conda deactivate
conda env remove --name session3_env
You can recreate the environment on the same machine you deleted it from
conda create --name session3_env --file ~/.conda/envs/session3_env_spec_file.txt
Running the Session 3 Tutorial with Your New Environment
During the Session 3 Tutorial we ran the tutorial in a container that already contained all the necessary software but use of a container wasn’t totally necessary. The session3_env Conda environment we have created also contains all the software needed to run it.
To run the Session3 Tutorial using your new session3_env Conda environment, navigate in JupyterLab to where you saved the Session 3 .ipynb file. If you didn’t participate in Session 3, go to the workshop website tutorial page for instructions on how to download the .ipynb file.
Once you have the Session 3 Jupyter notebook open, on the top right of the notebook you should be able to click to select a Kernel. In the Select Kernel window that pops up choose you session3_env from the dropdown list and then click Select. You are now running the Jupyter notebook in your session3_env Conda environment!
Containers (Docker and Singularity)
Why Use Containers?
Containers are a virtualized run-time environment- they are like handing someone the computer you used to execute your code. They allow you to bundle your software and its dependencies (including operating-system specific dependencies) in one tidy package that will run a specific isolated software environment on an operating system (e.g, Windos, Max, Linux flavors).
If you’ve ever written code on one machine and then tried to run it without success on a different machine, then you could benefit from using containers. As you can imagine, using containers to make your code portable means that sharing code between collaborators is very simple. And portable code makes it much easier for others to reproduce your results.
This tutorial focuses on creating container images with Docker and running the images on the Ceres HPC with Singularity. You can also create container images with Singularity and store/version them on Singularity-hub but we have chosen to feature a mostly Docker workflow because Docker container images are currently much more common (outside the HPC ecosystem) than Singularity. For more detailed information about singularity on Ceres, see this SCINet page.
Container Terminology
DOCKER - open source containerization software that you use at the command line to build and run container images. You can use docker on your laptop but it is not available on Ceres due to security concerns.
SINGULARITY - like Docker, Singularity is open source containerization software that you use at the command line to build and run container images. Due to security concerns, Singularity is what Ceres HPC users must use to run container images. At the time of this writing, Ceres has Singularity version 3.5.3.
DOCKER HUB - an online repository hosting service where you can archive your container images and find/share container images. Docker Hub is like GitHub for your Docker images.
SINGULARITY HUB - an online repository hosting service where you can archive your container images and find/share container images. Singularity Hub is like GitHub for your Singularity images.
CONTAINER - a virtualized run-time environment where users can isolate applications from the underlying system. A container is created by running an image. On the Ceres HPC, we create containers using Singularity software to run Docker images.
CONTAINER IMAGE - or just image for short. A read-only file that contains a collection of files such as source code, libraries, dependencies, tools, and other files needed to run a container. The steps in producing an image are added in layers. In this tutorial, we are working with Docker and Singularity images. When an image is run it becomes a container.
DOCKERFILE - a script of instructions that define how to build a specific Docker image
SINGULARITY DEFINITION FILE - Singularity’s equivalent of a dockerfile
Finding an Existing Docker Image
Several organizations maintain excellent data science container stacks:
- Jupyter: (Interactive computing with Jupyter, focusing on Python and R): Documentation - GitHub - Docker Hub
- Rocker: (Interactive computing with RStudio, focusing on R): Documentation - GitHub - Docker Hub
- RapidsAI: (Interactive computing with Jupyter+Python and GPUs): Documentation - GitHub - Docker Hub
If you’ve downloaded Docker on your laptop, you can search Docker Hub at the command line with docker search your-search-term, very similar to how you can search for packages with Conda. In addition, you can simply do a key-word on Docker Hub website of the +100,000 public containers. One limitation of Singularity on Ceres is that singularity search won’t search Docker Hub- it can only search it’s own singularity-hub registry.
Using a Docker Image on Ceres with Singularity
I.e., running a singularity container from a Docker image. On Ceres we must use singularity instead of docker (for security reasons) to download, build, and run container images. But we can still access/use any image on Docker Hub. The following will download docker image layers from Docker Hub, assemble them into a Singularity image, and run the Singularity container on Ceres.
Alpine is a popular (+1Billion Docker Hub pull requests) and lightweight linux OS. Here are are the steps to using this container on Ceres.
Step 1: Navigate to your home directory and download+build the container with:
navigate to HOME directory
cd $HOME
start an interactive session on a compute node
salloc
download and build a singularity container from dockerhub
singularity pull docker://alpine:latest
a container named alpine_latest.sif is in your folder.
ls
Step 2: Interact with the container.
shell (interactively enter) into the container with:
singularity shell alpine_latest.sif
Notice that the terminal has now changed (it should start with Singularity>). Lets see what OS we are using:
cat /etc/os-release
To leave the container, simply:
exit
Now the terminal has changed back to the standard Ceres compute node convention.
Step 3: Run a command inside the container from the host system.
Rather than the shell command, we will use the exec to run a command from within the container.
singularity exec alpine_latest.sif cat /etc/os-release
Bonus Step 4: Caching
Singularity caches the containers from Docker and Singularity Hub in your $HOME/.singularity folder. For instance, we can delete the alpine image we just made, and then simply shell into it using the “Docker Hub” url.
delete the container
rm alpine_latest.sif
it is no longer in the folder
ls
Now lets shell into the alpine image.
singularity shell docker://alpine:latest
Note that we did not need to download or rebuild the container. Singularity checks the local (cached) container against the container in the Hub repository. If they are the same, it will use the cached container. If the local container and the container in the Hub are different, it will download and rebuild the container. Now let’s exit the container and the Ceres compute node with:
exit the container
exit
exit the compute node
exit
Building a Docker Image
Option 1: Build an image on your local system and transfer to Ceres (not covered in this tutorial).
Option 2: Build an image with Docker Hub. There are multiple ways to accomplish this task. Here we discuss one approach to link GitHub to Docker Hub.
- Create GitHub and Docker Hub accounts.
- Create a GitHub repository, perhaps something like my_container.
- Within the my_container GitHub repository, make a file titled Dockerfile. Below is an example what a simple Dockerfile might look like:
FROM jupyter/datascience-notebook
LABEL maintainer="My Name <my.email@xxx.xxx>"
conda create --name session3_env python=3.7 numpy dask dask-jobqueue ipykernel
The above Dockerfile adds to the jupyter/datascience-notebook container. We first change the maintainer information to our information, then create a new conda environment named session3_env, and install python, numpy, dask, dask-jobqueue and the ipykernel.
Step 4. Create a new repository in Docker Hub and link it to the my_containers repository in GitHub.
Step 5. Trigger the image build in Docker Hub. Wait for the build to complete and then pull to Ceres.
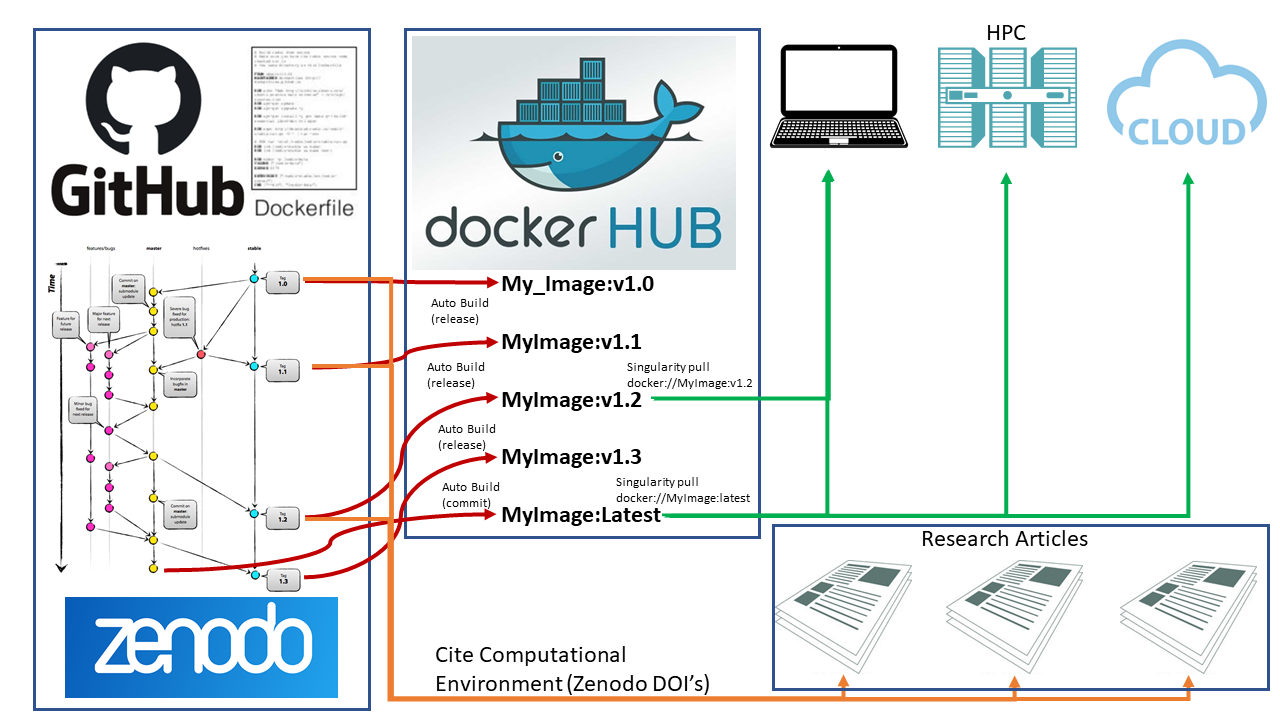
Example: GitHub/Zenodo + Docker Hub Setup
The tutorials for this workshop (SCINet Geospatial Workshop 2020) use an image titled data_science_im_rs_vSCINetGeoWS_2020.sif. The container was developed using GitHub (https://github.com/rmg55/container_stacks/tree/master/data_science_im_rs) and Docker Hub (https://hub.docker.com/r/rowangaffney/data_science_im_rs/tags). These repositories are set up to have some additional useful features:
- Docker Hub Auto-Build: Whenever the GitHub repository changes, a new image is automatically built on Docker Hub. This new image has the :latest tag.
- Capture GitHub Tags/Releases: When a release/tag is created in the GitHub repository, a new image is automatically built on Docker Hub with similiar tag (:vTAGNAME). For example, for this workshop, I created a release in the GitHub repository titled SCINetGeoWS_2020 (see: link to GitHub release). A subsequent image in Docker Hub was built (link equivalent Docker Hub Image). This image will not be over-written by subsequent changes in the GitHub repository (unlike the :latest image).
- Multiple Computing Environments: The container includes two conda environments (py_geo and r_geo) that focus on geospatial and machine learning software.
- Several IDEs: The container includes Jupyter, RStudio, and VSCode.
The above setup (with the addition of Zenodo linked to the GitHub repository) would support the workflow outlined in the below figure. The container images are archived/stored on Docker Hub while the code used to define and build the images is archived on GitHub and citable via a Zenodo DOI. This workflow allows for a consistent and reproducible computational environment from laptop/desktop to HPC to cloud computing systems.